Tutorial
Authors: Pierre-Henri Tournier - Frédéric Nataf - Pierre Jolivet
We recommend checking the slides version of this tutorial.
What is ffddm ?
ffddm implements a class of parallel solvers in FreeFEM: overlapping Schwarz domain decomposition methods
- ffddm provides a set of high-level macros and functions to
handle data distribution: distributed meshes and linear algebra
build DD preconditioners for your variational problems
solve your problem using preconditioned Krylov methods
ffddm implements scalable two level Schwarz methods, with a coarse space correction built either from a coarse mesh or a GenEO coarse space Ongoing research: approximate coarse solves and three level methods
Why Domain Decomposition Methods ?
How can we solve a large sparse linear system \(A u = b \in \mathbb{R}^n\) ?
Step 1: Decompose the mesh
See documentation
Build a collection of \(N\) overlapping sub-meshes \((Th_{i})_{i=1}^N\) from the global mesh \(Th\)
1ffddmbuildDmesh( prmesh , ThGlobal , comm )
mesh distributed over the MPI processes of communicator comm
initial mesh ThGlobal partitioned with metis by default
size of the overlap given by ffddmoverlap (default 1)
prmesh#Thi is the local mesh of the subdomain for each mpi process
1macro dimension 2// EOM // 2D or 3D
2
3include "ffddm.idp"
4
5mesh ThGlobal = square(100,100); // global mesh
6
7// Step 1: Decompose the mesh
8ffddmbuildDmesh( M , ThGlobal , mpiCommWorld )
9
10medit("Th"+mpirank, MThi);
Copy and paste this to a file ‘test.edp’ and run it:
1ff-mpirun -np 2 test.edp -wg
Step 2: Define your finite element
See documentation
1ffddmbuildDfespace( prfe , prmesh , scalar , def , init , Pk )
builds the local finite element spaces and associated distributed operators on top of the mesh decomposition prmesh
scalar: type of data for this finite element: real or complex
Pk: your type of finite element: P1, [P2,P2,P1], …
def, init: macros specifying how to define and initialize a Pk FE function
prfe#Vhi is the local FE space defined on prmesh#Thi for each mpi process
Example for P2 complex:
1macro def(u) u // EOM
2macro init(u) u // EOM
3ffddmbuildDfespace( FE, M, complex,
4 def, init, P2 )
Example for [P2,P2,P1] real:
1macro def(u) [u, u#B, u#C] // EOM
2macro init(u) [u, u, u] // EOM
3ffddmbuildDfespace( FE, M, real, def,
4 init, [P2,P2,P1] )
Distributed vectors and restriction operators
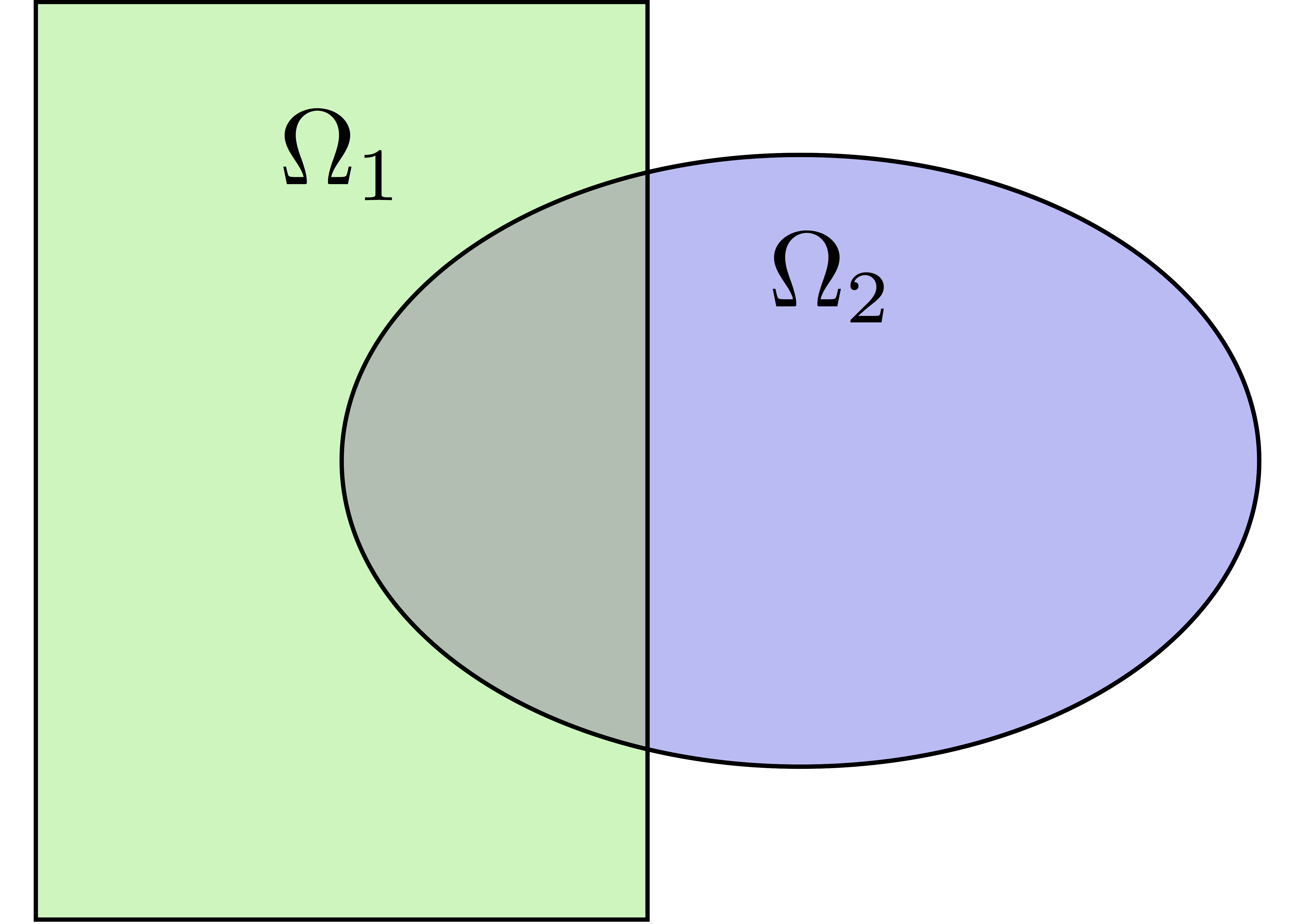
Natural decomposition of the set of d.o.f.’s \({\mathcal N}\) of \(Vh\) into the \(N\) subsets of d.o.f.’s \(({\mathcal N}_i)_{i=1}^N\) each associated with the local FE space \(Vh_i\)
but with duplications of the d.o.f.’s in the overlap
_Definition_ a distributed vector is a collection of local vectors \(({\mathbf V_i})_{1\le i\le N}\) so that the values on the duplicated d.o.f.’s are the same:
where \({\mathbf V}\) is the corresponding global vector and \(R_i\) is the restriction operator from \({\mathcal N}\) into \({\mathcal N}_i\)
Remark \(R_i^T\) is the extension operator: extension by \(0\) from \({\mathcal N}_i\) into \({\mathcal N}\)
Partition of unity
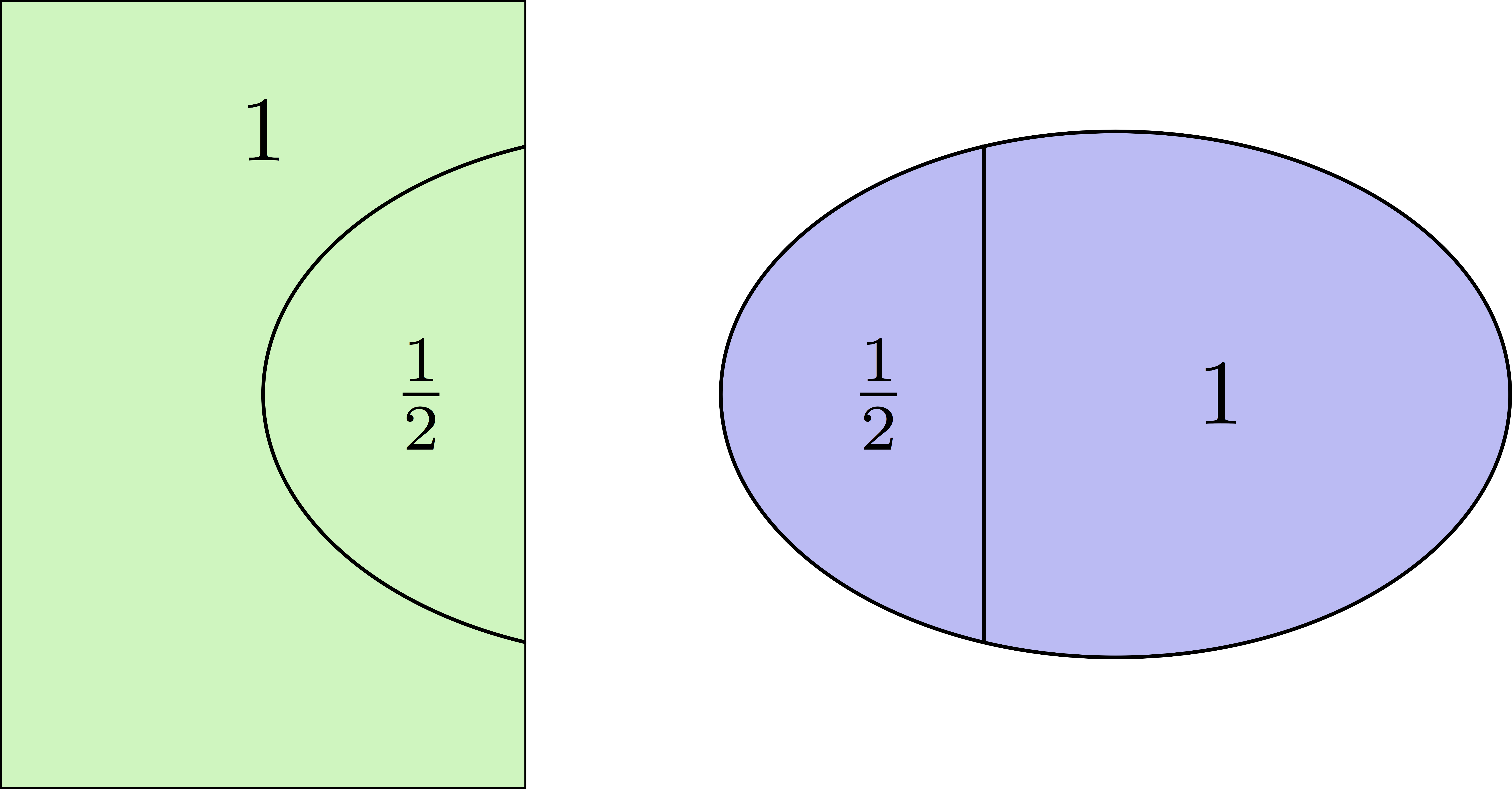
Duplicated unknowns coupled via a partition of unity:
\((D_i)_{1\le i \le N}\) are square diagonal matrices of size \(\#{\mathcal N}_i\)
Data exchange between neighbors
1func prfe#update(K[int] vi, bool scale)
synchronizes local vectors \({\mathbf V}_i\) between subdomains \(\Rightarrow\) exchange the values of \(mathbf{V}_i\) shared with neighbors in the overlap region
where \(\mathcal{O}(i)\) is the set of neighbors of subdomain $i$. Exchange operators \(R_i\,R_j^T\) correspond to neighbor-to-neighbor MPI communications
1FEupdate(vi, false);
1FEupdate(vi, true);
1macro dimension 2// EOM // 2D or 3D
2
3include "ffddm.idp"
4
5mesh ThGlobal = square(100,100); // global mesh
6
7// Step 1: Decompose the mesh
8ffddmbuildDmesh( M , ThGlobal , mpiCommWorld )
9
10// Step 2: Define your finite element
11macro def(u) u // EOM
12macro init(u) u // EOM
13ffddmbuildDfespace( FE , M , real , def , init , P2 )
14
15FEVhi vi = x;
16medit("v"+mpirank, MThi, vi);
17
18vi[] = FEDk[mpirank];
19medit("D"+mpirank, MThi, vi);
20
21vi = 1;
22FEupdate(vi[],true);
23ffddmplot(FE,vi,"1")
24
25FEupdate(vi[],false);
26ffddmplot(FE,vi,"multiplicity")
Step 3: Define your problem
See documentation
1ffddmsetupOperator( pr , prfe , Varf )
builds the distributed operator associated to your variational form on top of the distributed FE prfe
Varf is a macro defining your abstract variational form
1macro Varf(varfName, meshName, VhName)
2 varf varfName(u,v) = int2d(meshName)(grad(u)'* grad(v))
3 + int2d(meshName)(f*v) + on(1, u = 0); // EOM
\(\Rightarrow\) assemble local ‘Dirichlet’ matrices \(A_i = R_i A R_i^T\)
Warning
only true because \(D_i R_i A = D_i A_i R_i\) due to the fact that \(D_i\) vanishes at the interface !!
pr#A applies \(A\) to a distributed vector: \({\mathbf U}_i \leftarrow R_i \sum_{j=1}^N R_j^T D_j A_j {\mathbf V}_j\)
\(\Rightarrow\) multiply by \(A_i\) + prfe#update
1macro dimension 2// EOM // 2D or 3D
2
3include "ffddm.idp"
4
5mesh ThGlobal = square(100,100); // global mesh
6
7// Step 1: Decompose the mesh
8ffddmbuildDmesh( M , ThGlobal , mpiCommWorld )
9
10// Step 2: Define your finite element
11macro def(u) u // EOM
12macro init(u) u // EOM
13ffddmbuildDfespace( FE , M , real , def , init , P2 )
14
15// Step 3: Define your problem
16macro grad(u) [dx(u), dy(u)] // EOM
17macro Varf(varfName, meshName, VhName)
18 varf varfName(u,v) = int2d(meshName)(grad(u)'* grad(v))
19 + int2d(meshName)(1*v) + on(1, u = 0); // EOM
20ffddmsetupOperator( PB , FE , Varf )
21
22FEVhi ui, bi;
23ffddmbuildrhs( PB , Varf , bi[] )
24
25ui[] = PBA(bi[]);
26ffddmplot(FE, ui, "A*b")
Summary so far: translating your sequential FreeFEM script
Step 1: Decompose the mesh
See documentation
1mesh Th = square(100,100);
1mesh Th = square(100,100);
2ffddmbuildDmesh(M, Th, mpiCommWorld)
Step 2: Define your finite element
See documentation
1fespace Vh(Th, P1);
1macro def(u) u // EOM
2macro init(u) u // EOM
3ffddmbuildDfespace(FE, M, real, def, init, P1)
Step 3: Define your problem
See documentation
1varf Pb(u, v) = ...
2matrix A = Pb(Vh, Vh);
1macro Varf(varfName, meshName, VhName)
2 varf varfName(u,v) = ... // EOM
3ffddmsetupOperator(PB, FE, Varf)
Solve the linear system
See documentation
1u[] = A^-1 * b[];
1ui[] = PBdirectsolve(bi[]);
Solve the linear system with the parallel direct solver MUMPS
See documentation
1func K[int] pr#directsolve(K[int]& bi)
We have \(A\) and \(b\) in distributed form, we can solve the linear system \(A u = b\) using the parallel direct solver MUMPS
1// Solve the problem using the direct parallel solver MUMPS
2ui[] = PBdirectsolve(bi[]);
3ffddmplot(FE, ui, "u")
Step 4: Define the one level DD preconditioner
See documentation
1ffddmsetupPrecond( pr , VarfPrec )
builds the one level preconditioner for problem pr.
By default it is the Restricted Additive Schwarz (RAS) preconditioner:
_Setup step_: compute the \(LU\) (or \(L D L^T\)) factorization of local matrices \(A_i\)
pr#PREC1 applies \(M^{-1}_1\) to a distributed vector: \({\mathbf U}_i \leftarrow R_i \sum_{j=1}^N R_j^T D_j A_j^{-1} {\mathbf V}_i\)
\(\Rightarrow\) apply \(A_i^{-1}\) (forward/backward substitutions) + prfe#update
Step 5: Solve the linear system with preconditioned GMRES
See documentation
1func K[int] pr#fGMRES(K[int]& x0i, K[int]& bi, real eps, int itmax, string sp)
solves the linear system with flexible GMRES with DD preconditioner \(M^{-1}\)
x0i: initial guess
bi: right-hand side
eps: relative tolerance
itmax: maximum number of iterations
sp: “left” or “right” preconditioning
left preconditioning
solve \(M^{-1} A x = M^{-1} b\)
right preconditioning
solve \(A M^{-1} y = b\)
\(\Rightarrow x = M^{-1} y\)
1macro dimension 2// EOM // 2D or 3D
2include "ffddm.idp"
3
4mesh ThGlobal = square(100,100); // global mesh
5// Step 1: Decompose the mesh
6ffddmbuildDmesh( M , ThGlobal , mpiCommWorld )
7// Step 2: Define your finite element
8macro def(u) u // EOM
9macro init(u) u // EOM
10ffddmbuildDfespace( FE , M , real , def , init , P2 )
11// Step 3: Define your problem
12macro grad(u) [dx(u), dy(u)] // EOM
13macro Varf(varfName, meshName, VhName)
14 varf varfName(u,v) = int2d(meshName)(grad(u)'* grad(v))
15 + int2d(meshName)(1*v) + on(1, u = 0); // EOM
16ffddmsetupOperator( PB , FE , Varf )
17
18FEVhi ui, bi;
19ffddmbuildrhs( PB , Varf , bi[] )
20
21// Step 4: Define the one level DD preconditioner
22ffddmsetupPrecond( PB , Varf )
23
24// Step 5: Solve the linear system with GMRES
25FEVhi x0i = 0;
26ui[] = PBfGMRES(x0i[], bi[], 1.e-6, 200, "right");
27
28ffddmplot(FE, ui, "u")
29PBwritesummary
Define a two level DD preconditioner
See documentation
Goal improve scalability of the one level method
\(\Rightarrow\) enrich the one level preconditioner with a coarse problem coupling all subdomains
Main ingredient is a rectangular matrix \(\color{red}{Z}\) of size \(n \times n_c,\,\) where \(n_c \ll n\) \(\color{red}{Z}\) is the coarse space matrix
The coarse space operator \(E = \color{red}{Z}^T A \color{red}{Z}\) is a square matrix of size \(n_c \times n_c\)
The simplest way to enrich the one level preconditioner is through the additive coarse correction formula:
How to choose \(\color{red}{Z}\) ?
Build the GenEO coarse space
See documentation
1ffddmgeneosetup( pr , Varf )
The GenEO method builds a robust coarse space for highly heterogeneous or anisotropic SPD problems
\(\Rightarrow\) solve a local generalized eigenvalue problem in each subdomain
with \(A_i^{\text{Neu}}\) the local Neumann matrices built from Varf (same Varf as Step 3)
The GenEO coarse space is \(\color{red}{Z} = (R_i^T D_i V_{i,k})^{i=1,...,N}_{\lambda_{i,k} \ge \color{blue}{\tau}}\) The eigenvectors \(V_{i,k}\) selected to enter the coarse space correspond to eigenvalues \(\lambda_{i,k} \ge \color{blue}{\tau}\), where \(\color{blue}{\tau}\) is a threshold parameter
Theorem the spectrum of the preconditioned operator lies in the interval \([\displaystyle \frac{1}{1+k_1 \color{blue}{\tau}} , k_0 ]\) where \(k_0 - 1\) is the # of neighbors and \(k_1\) is the multiplicity of intersections \(\Rightarrow\) \(k_0\) and \(k_1\) do not depend on \(N\) nor on the PDE
1macro dimension 2// EOM // 2D or 3D
2include "ffddm.idp"
3
4mesh ThGlobal = square(100,100); // global mesh
5// Step 1: Decompose the mesh
6ffddmbuildDmesh( M , ThGlobal , mpiCommWorld )
7// Step 2: Define your finite element
8macro def(u) u // EOM
9macro init(u) u // EOM
10ffddmbuildDfespace( FE , M , real , def , init , P2 )
11// Step 3: Define your problem
12macro grad(u) [dx(u), dy(u)] // EOM
13macro Varf(varfName, meshName, VhName)
14 varf varfName(u,v) = int2d(meshName)(grad(u)'* grad(v))
15 + int2d(meshName)(1*v) + on(1, u = 0); // EOM
16ffddmsetupOperator( PB , FE , Varf )
17
18FEVhi ui, bi;
19ffddmbuildrhs( PB , Varf , bi[] )
20
21// Step 4: Define the one level DD preconditioner
22ffddmsetupPrecond( PB , Varf )
23
24// Build the GenEO coarse space
25ffddmgeneosetup( PB , Varf )
26
27// Step 5: Solve the linear system with GMRES
28FEVhi x0i = 0;
29ui[] = PBfGMRES(x0i[], bi[], 1.e-6, 200, "right");
Build the coarse space from a coarse mesh
See documentation
1ffddmcoarsemeshsetup( pr , Thc , VarfEprec , VarfAprec )
For non SPD problems, an alternative is to build the coarse space by discretizing the PDE on a coarser mesh Thc
\(Z\) will be the interpolation matrix from the coarse FE space \({Vh}_c\) to the original FE space \(Vh\)
\(\Rightarrow E=\color{red}{Z}^{T} A \color{red}{Z}\) is the matrix of the problem discretized on the coarse mesh
The variational problem to be discretized on Thc is given by macro VarfEprec
VarfEprec can differ from the original Varf of the problem
Example: added absorption for wave propagation problems
Similarly, VarfAprec specifies the global operator involved in multiplicative coarse correction formulas. It defaults to \(A\) if VarfAprec is not defined
1macro dimension 2// EOM // 2D or 3D
2include "ffddm.idp"
3
4mesh ThGlobal = square(100,100); // global mesh
5// Step 1: Decompose the mesh
6ffddmbuildDmesh( M , ThGlobal , mpiCommWorld )
7// Step 2: Define your finite element
8macro def(u) u // EOM
9macro init(u) u // EOM
10ffddmbuildDfespace( FE , M , real , def , init , P2 )
11// Step 3: Define your problem
12macro grad(u) [dx(u), dy(u)] // EOM
13macro Varf(varfName, meshName, VhName)
14 varf varfName(u,v) = int2d(meshName)(grad(u)'* grad(v))
15 + int2d(meshName)(1*v) + on(1, u = 0); // EOM
16ffddmsetupOperator( PB , FE , Varf )
17
18FEVhi ui, bi;
19ffddmbuildrhs( PB , Varf , bi[] )
20
21// Step 4: Define the one level DD preconditioner
22ffddmsetupPrecond( PB , Varf )
23
24// Build the coarse space from a coarse mesh
25mesh Thc = square(10,10);
26ffddmcoarsemeshsetup( PB , Thc , Varf , null )
27
28// Step 5: Solve the linear system with GMRES
29FEVhi x0i = 0;
30ui[] = PBfGMRES(x0i[], bi[], 1.e-6, 200, "right");
Use HPDDM within ffddm
See documentation
ffddm allows you to use HPDDM to solve your problem, effectively replacing the ffddm implementation of all parallel linear algebra computations
\(\Rightarrow\) define your problem with ffddm, solve it with HPDDM
\(\Rightarrow\) ffddm acts as a finite element interface for HPDDM
You can use HPDDM features unavailable in ffddm such as advanced Krylov subspace methods implementing block and recycling techniques
To switch to HPDDM, simply define the macro pr#withhpddm before using ffddmsetupOperator (Step 3). You can then pass HPDDM options with command-line arguments or directly to the underlying HPDDM operator. Options need to be prefixed by the operator prefix:
1macro PBwithhpddm()1 // EOM
2ffddmsetupOperator( PB , FE , Varf )
3set(PBhpddmOP,sparams="-hpddm_PB_krylov_method gcrodr -hpddm_PB_recycle 10");
Or, define pr#withhpddmkrylov to use HPDDM only for the Krylov method
Example here: Helmholtz problem with multiple rhs solved with Block GMRES
Some results: Heterogeneous 3D elasticity with GenEO
Heterogeneous 3D linear elasticity equation discretized with P2 FE solved on 4096 MPI processes \(n\approx\) 262 million
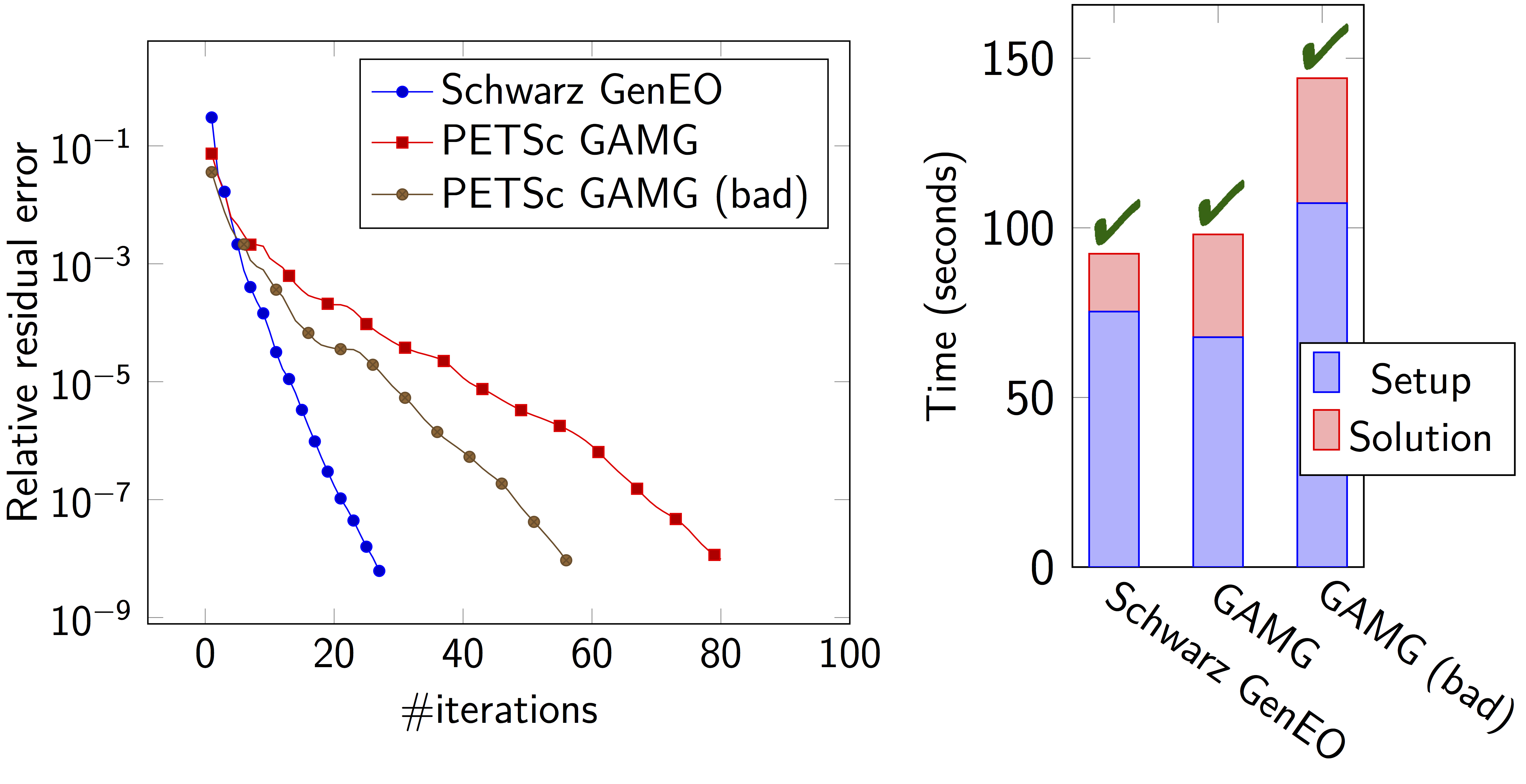
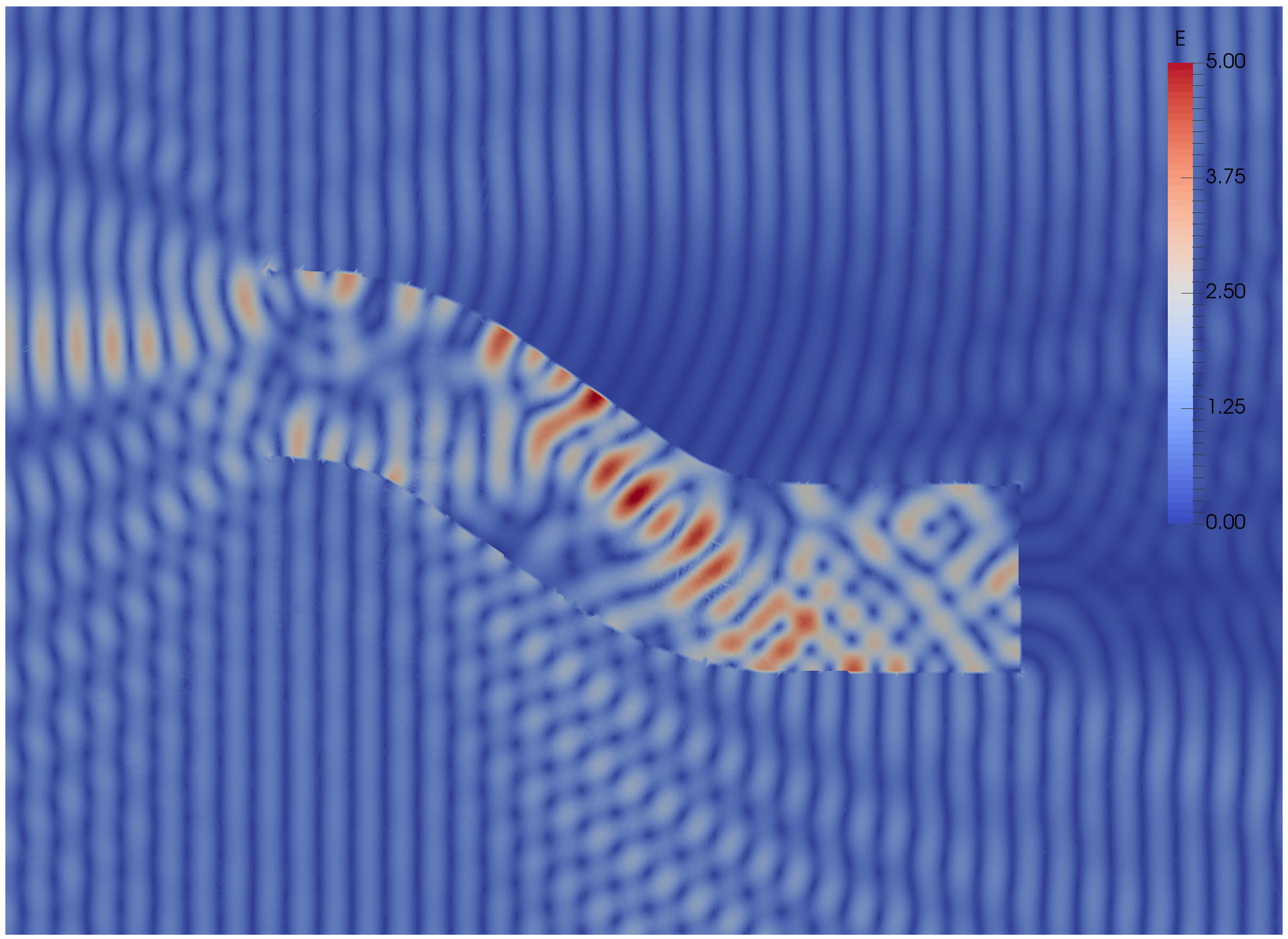
Some results: 2-level DD for Maxwell equations, scattering from the COBRA cavity
f = 10 GHz
f = 16 GHz
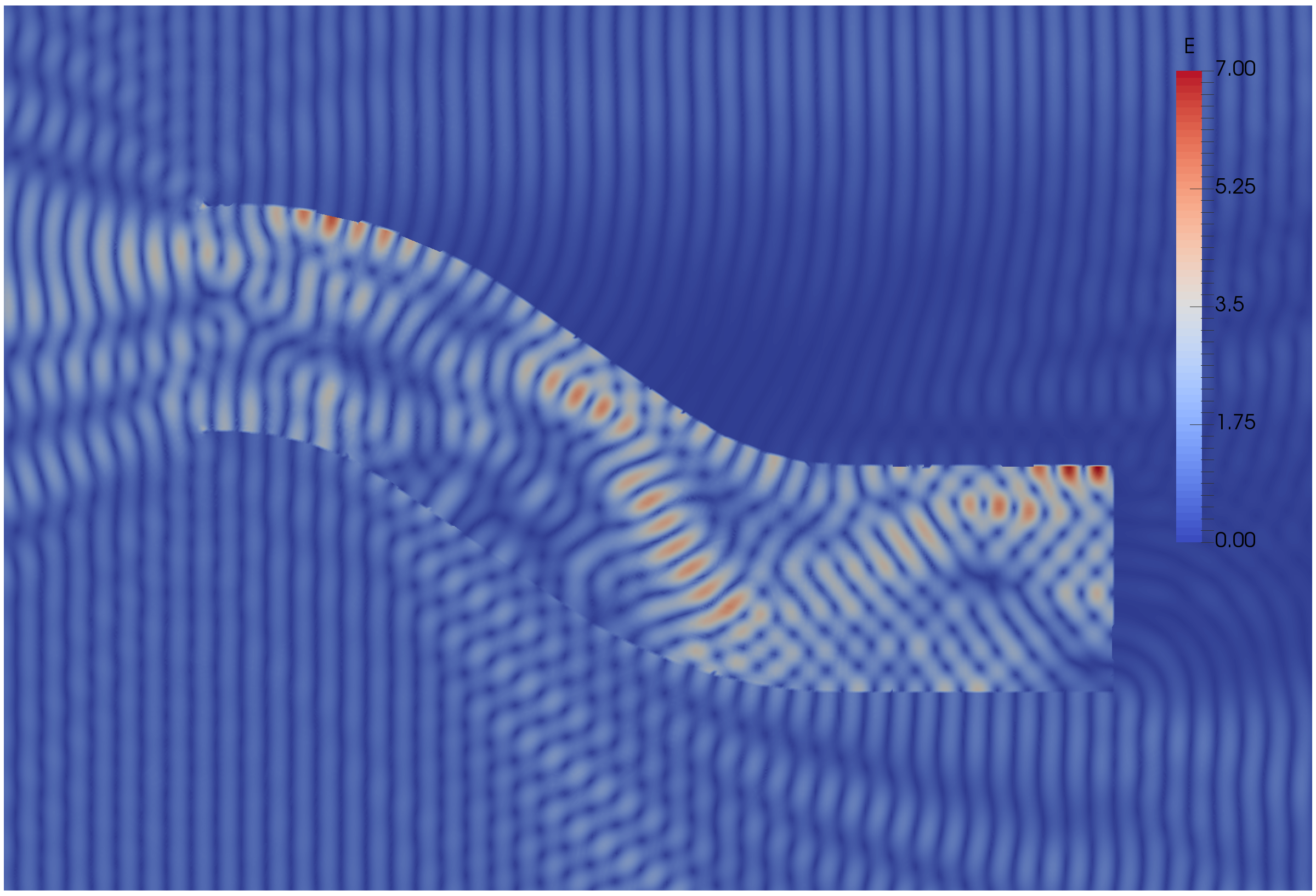
Some results: 2-level DD for Maxwell equations, scattering from the COBRA cavity
order 2 Nedelec edge FE
fine mesh: 10 points per wavelength
coarse mesh: 3.33 points per wavelength
two level ORAS preconditioner with added absorption
f = 10 GHz: \(n\approx\) 107 million, \(n_c \approx\) 4 million
f = 16 GHz: \(n\approx\) 198 million, \(n_c \approx\) 7.4 million
\(\rightarrow\) coarse problem too large for a direct solver \(\Rightarrow\) inexact coarse solve: GMRES + one level ORAS preconditioner
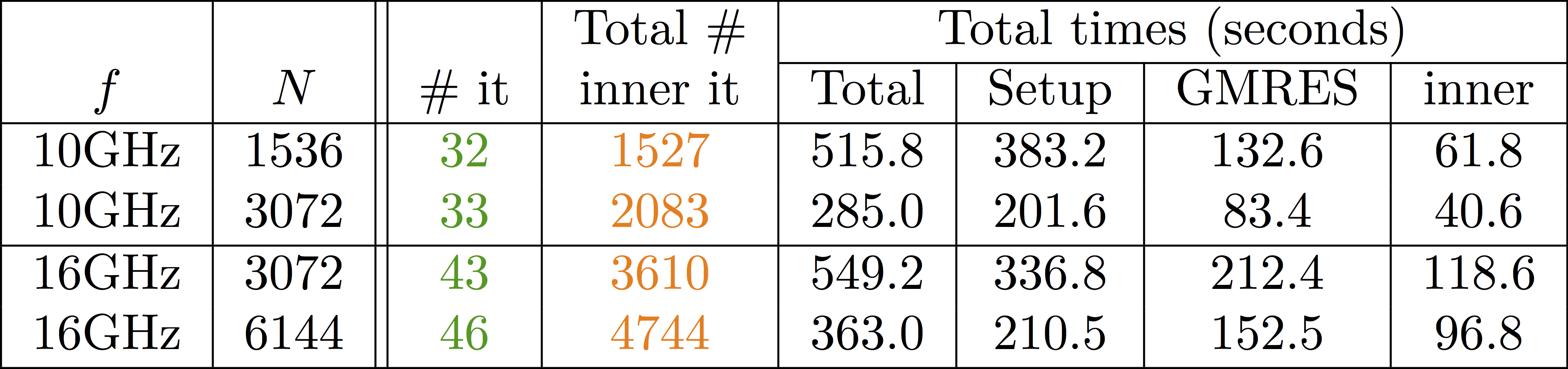
speedup of 1.81 from 1536 to 3072 cores at 10GHz
1.51 from 3072 to 6144 cores at 16GHz
You can find the script here