class: center, middle, inverse .title[ffddm] # Short tutorial .footnote1[Pierre-Henri Tournier Frédéric Nataf Pierre Jolivet] .footnote2[tournier@ljll.math.upmc.fr nataf@ljll.math.upmc.fr pierre@joliv.et] --- class: left, top ## [What is **ffddm** ?](../../documentation/ffddm/index.html#ffddm) - **ffddm** implements a class of parallel solvers in *FreeFEM*: *overlapping Schwarz domain decomposition methods* - The entire **ffddm** framework is written in the *FreeFEM* language **ffddm** aims at simplifying the use of parallel solvers in *FreeFEM* You can find the **ffddm** scripts [here](https://github.com/FreeFem/FreeFem-sources/blob/develop/idp) ('ffddm*.idp' files) and examples [here](https://github.com/FreeFem/FreeFem-sources/blob/develop/examples/ffddm) - **ffddm** provides a set of high-level macros and functions to - handle data distribution: distributed meshes and linear algebra - build DD preconditioners for your variational problems - solve your problem using preconditioned Krylov methods - **ffddm** implements scalable two level Schwarz methods, with a coarse space correction built either from a coarse mesh or a [GenEO](https://link.springer.com/article/10.1007%2Fs00211-013-0576-y#page-1) coarse space *Ongoing research*: approximate coarse solves and three level methods - **ffddm** can also act as a wrapper for the [**HPDDM**](https://github.com/hpddm/hpddm) library **HPDDM** is an efficient C++11 implementation of various domain decomposition methods and Krylov subspace algorithms with advanced block and recycling techniques More details on how to use **HPDDM** within **ffddm** [here](../../documentation/ffddm/documentation.html#using-hpddm-within-ffddm) --- ## Why Domain Decomposition Methods ? How can we solve a large sparse linear system $A u = b \in \mathbb{R}^n$ ? 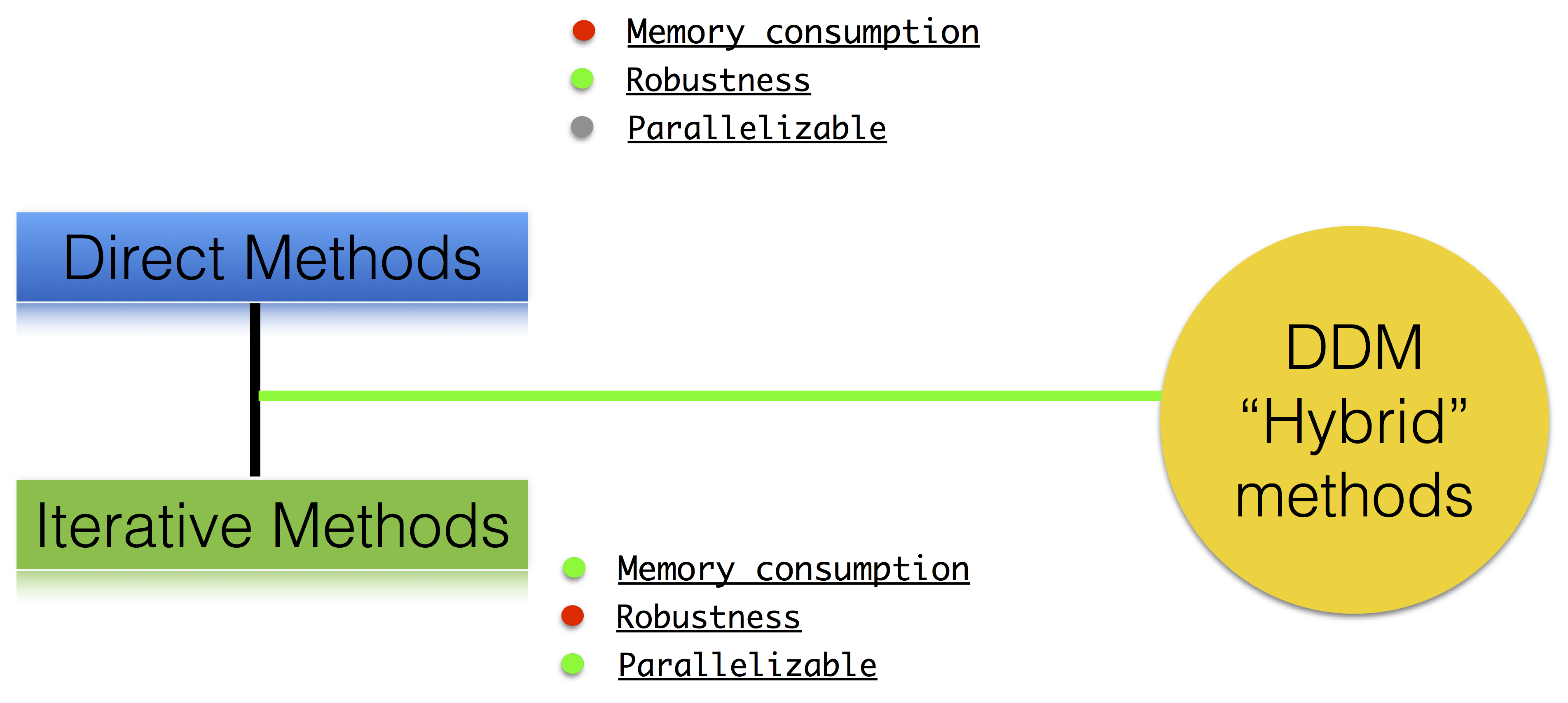 --- ## [Step 1: Decompose the mesh](../../documentation/ffddm/documentation.html#overlapping-mesh-decomposition) Build a collection of $N$ overlapping sub-meshes $(Th\_{i})\_{i=1}^N$ from the global mesh $Th$ <div style="overflow: hidden; display: inline-block;"> <img src="../../_images/domain1.png" width = 35% style="float:left" /> <img src="../../_images/domain2.png" width = 35% style="float:right"/> </div> ```cpp ffddmbuildDmesh( prmesh , ThGlobal , comm ) ``` - mesh distributed over the mpi processes of communicator **comm** - initial mesh **ThGlobal** partitioned with *metis* by default - size of the overlap given by [ffddmoverlap](../../documentation/ffddm/parameters.html#global-parameters) (default 1) `prmesh#Thi` is the local mesh of the subdomain for each mpi process --- ```cpp macro dimension 2// EOM // 2D or 3D include "ffddm.idp" mesh ThGlobal = square(100,100); // global mesh *// Step 1: Decompose the mesh *ffddmbuildDmesh( M , ThGlobal , mpiCommWorld ) ``` ```cpp medit("Th"+mpirank, MThi); ``` Copy and paste this to a file 'test.edp' and run it: ```console $ ff-mpirun -np 2 test.edp -wg ``` --- ## [Step 2: Define your finite element](../../documentation/ffddm/documentation.html#local-finite-element-spaces) ```cpp ffddmbuildDfespace( prfe , prmesh , scalar , def , init , Pk ) ``` builds the local finite element spaces and associated distributed operators on top of the mesh decomposition **prmesh** - **scalar**: type of data for this finite element: *real* or *complex* - **Pk**: your type of finite element: P1, [P2,P2,P1], ... - **def**, **init**: macros specifying how to define and initialize a **Pk** FE function `prfe#Vhi` is the local FE space defined on `prmesh#Thi` for each mpi process .pull-left[ example for P2 *complex*: ```cpp macro def(u) u // EOM macro init(u) u // EOM ffddmbuildDfespace( FE, M, complex, def, init, P2 ) ``` ] .pull-right[ example for [P2,P2,P1] *real*: ```cpp macro def(u) [u, u#B, u#C] // EOM macro init(u) [u, u, u] // EOM ffddmbuildDfespace( FE, M, real, def, init, [P2,P2,P1] ) ``` ] --- ## [Step 2: Define your finite element](../../documentation/ffddm/documentation.html#local-finite-element-spaces) ### Distributed vectors and restriction operators <div> <img src="../../_images/domain2.png" width = 35% style="float:right"/> </div> Natural decomposition of the set of d.o.f.'s ${\mathcal N}$ of $Vh$ into the $N$ subsets of d.o.f.'s $({\mathcal N}\_i)\_{i=1}^N$ each associated with the local FE space $Vh\_i$ $${\mathcal N} = \cup\_{i=1}^N {\mathcal N}\_i\,, $$ but with duplications of the d.o.f.'s in the overlap **_Definition_** a *distributed vector* is a collection of local vectors $({\mathbf V\_i})\_{1\le i\le N}$ so that the values on the duplicated d.o.f.'s are the same: $$ {\mathbf V}\_i = R\_i\,{\mathbf V}, \quad i = 1, ..., N $$ where ${\mathbf V}$ is the corresponding global vector and $R\_i$ is the *restriction operator* from ${\mathcal N}$ into ${\mathcal N}\_i$ **_Remark_** $R\_i^T$ is the *extension operator*: extension by $0$ from ${\mathcal N}\_i$ into ${\mathcal N}$ --- ## [Step 2: Define your finite element](../../documentation/ffddm/documentation.html#local-finite-element-spaces) ### Partition of unity <div style="overflow: hidden; display: inline-block;"> <div> <img src="../../_images/domain3.png" width = 43% style="float:right"/> </div> Duplicated unknowns coupled via a *partition of unity*: $$ I = \sum\_{i = 1}^N R\_i^T D\_i R\_i $$ </div> $(D\_i)\_{1\le i \le N}$ are square diagonal matrices of size #${\mathcal N}\_i$ $$ {\mathbf V} = \sum\_{i = 1}^N R\_i^T D\_i R\_i {\mathbf V} = \sum\_{i = 1}^N R\_i^T D\_i {\mathbf V\_i} $$ --- ## [Step 2: Define your finite element](../../documentation/ffddm/documentation.html#local-finite-element-spaces) ### Data exchange between neighbors ```cpp func prfe#update(K[int] vi, bool scale) ``` synchronizes local vectors ${\mathbf V}\_i$ between subdomains $\Rightarrow$ exchange the values of ${\mathbf V}\_i$ shared with neighbors in the overlap region $$ {\mathbf V}\_i \leftarrow R\_i \left( \sum\_{j=1}^N R\_j^T D\_j {\mathbf V}\_j \right) = D\_i {\mathbf V}\_i + \sum\_{j\in \mathcal{O}(i)} R\_i\,R\_j^T\,D\_j {\mathbf V}\_j $$ where $\mathcal{O}(i)$ is the set of neighbors of subdomain $i$. Exchange operators $R_i\,R_j^T$ correspond to neighbor-to-neighbor MPI communications .pull-left[ ```cpp FEupdate(vi, false); ``` $$ {\mathbf V}\_i \leftarrow R\_i \left( \sum\_{j=1}^N R\_j^T {\mathbf V}\_j \right) $$ ] .pull-right[ ```cpp FEupdate(vi, true); ``` $$ {\mathbf V}\_i \leftarrow R\_i \left( \sum\_{j=1}^N R\_j^T D\_j {\mathbf V}\_j \right) $$ ] --- ```cpp macro dimension 2// EOM // 2D or 3D include "ffddm.idp" mesh ThGlobal = square(100,100); // global mesh // Step 1: Decompose the mesh ffddmbuildDmesh( M , ThGlobal , mpiCommWorld ) *// Step 2: Define your finite element *macro def(u) u // EOM *macro init(u) u // EOM *ffddmbuildDfespace( FE , M , real , def , init , P2 ) ``` ```cpp FEVhi vi = y; medit("v"+mpirank, MThi, vi); vi[] = FEDk[mpirank]; medit("D"+mpirank, MThi, vi); vi = 1; FEupdate(vi[],true); ffddmplot(FE,vi,"1") FEupdate(vi[],false); ffddmplot(FE,vi,"multiplicity") ``` --- ## [Step 3: Define your problem](../../documentation/ffddm/documentation.html#define-the-problem-to-solve) ```cpp ffddmsetupOperator( pr , prfe , Varf ) ``` builds the distributed operator associated to your variational form on top of the distributed FE **prfe** **Varf** is a macro defining your abstract variational form ```cpp macro Varf(varfName, meshName, VhName) varf varfName(u,v) = int2d(meshName)(grad(u)'* grad(v)) + int2d(meshName)(f*v) + on(1, u = 0); // EOM ``` $\Rightarrow$ assemble local 'Dirichlet' matrices $A\_i = R\_i A R\_i^T$ <div style="overflow: hidden;"> .pull-left31[ $$A = \sum\_{i=1}^N R\_i^T D\_i A\_i R\_i$$ ] .pull-right65[ **Warning** only true because $D\_i R\_i A = D\_i A\_i R\_i$ due to the fact that $D\_i$ vanishes at the interface **!!** ] </div> `pr#A` applies $A$ to a distributed vector: ${\mathbf U}\_i \leftarrow R\_i \sum\_{j=1}^N R\_j^T D\_j A\_j {\mathbf V}\_j$ $\Rightarrow$ multiply by $A\_i$ + `prfe#update` --- ```cpp macro dimension 2// EOM // 2D or 3D include "ffddm.idp" mesh ThGlobal = square(100,100); // global mesh // Step 1: Decompose the mesh ffddmbuildDmesh( M , ThGlobal , mpiCommWorld ) // Step 2: Define your finite element macro def(u) u // EOM macro init(u) u // EOM ffddmbuildDfespace( FE , M , real , def , init , P2 ) *// Step 3: Define your problem *macro grad(u) [dx(u), dy(u)] // EOM *macro Varf(varfName, meshName, VhName) * varf varfName(u,v) = int2d(meshName)(grad(u)'* grad(v)) * + int2d(meshName)(1*v) + on(1, u = 0); // EOM *ffddmsetupOperator( PB , FE , Varf ) * *FEVhi ui, bi; *ffddmbuildrhs( PB , Varf , bi[] ) ``` ```cpp ui[] = PBA(bi[]); ffddmplot(FE, ui, "A*b") ``` --- ## Summary so far: translating your sequential *FreeFEM* script <div style="overflow: hidden;"> .pull-left38[ ### [Step 1: Decompose the mesh](../../documentation/ffddm/documentation.html#overlapping-mesh-decomposition) ```ff1 mesh Th = square(100,100); ``` ] .pull-right58[ ```ff1 mesh Th = square(100,100); ffddmbuildDmesh(M, Th, mpiCommWorld) ``` ] </div> <div style="overflow: hidden;"> .pull-left38[ ### [Step 2: Define your finite element](../../documentation/ffddm/documentation.html#local-finite-element-spaces) ```ff1 fespace Vh(Th, P1); ``` ] .pull-right58[ ```ff1 macro def(u) u // EOM macro init(u) u // EOM ffddmbuildDfespace(FE, M, real, def, init, P1) ``` ] </div> <div style="overflow: hidden;"> .pull-left38[ ### [Step 3: Define your problem](../../documentation/ffddm/documentation.html#define-the-problem-to-solve) ```ff1 varf Pb(u, v) = ... matrix A = Pb(Vh, Vh); ``` ] .pull-right58[ ```ff1 macro Varf(varfName, meshName, VhName) varf varfName(u,v) = ... // EOM ffddmsetupOperator(PB, FE, Varf) ``` ] </div> <!-- <div style="overflow: hidden;"> .pull-left38[ ### [Solve the linear system](documentation/#define-the-problem-to-solve) ```ff1 u[] = A^-1 * b[]; ``` ] .pull-right58[ ```ff1 ui[] = PBdirectsolve(bi[]); ``` ] </div> --> --- ## [Solve the linear system with the parallel direct solver *MUMPS*](../../documentation/ffddm/documentation.html#define-the-problem-to-solve) ```cpp func K[int] pr#directsolve(K[int]& bi) ``` We have $A$ and $b$ in distributed form, we can solve the linear system $A u = b$ using the parallel direct solver *MUMPS* ```cpp // Solve the problem using the direct parallel solver MUMPS ui[] = PBdirectsolve(bi[]); ffddmplot(FE, ui, "u") ``` --- ## [Step 4: Define the one level DD preconditioner](../../documentation/ffddm/documentation.html#one-level-preconditioners) ```cpp ffddmsetupPrecond( pr , VarfPrec ) ``` builds the one level preconditioner for problem **pr**. By default it is the *Restricted Additive Schwarz (RAS)* preconditioner: $$ M^{-1}\_1 = M^{-1}\_{\text{RAS}} = \sum\_{i=1}^N R\_i^T D\_i A\_i^{-1} R\_i \quad \text{with}\; A\_i = R\_i A R\_i^T $$ **_Setup step_**: compute the $LU$ (or $L D L^T$) factorization of local matrices $A\_i$ `pr#PREC1` applies $M^{-1}\_1$ to a distributed vector: ${\mathbf U}\_i \leftarrow R\_i \sum\_{j=1}^N R\_j^T D\_j A\_j^{-1} {\mathbf V}\_i$ $\Rightarrow$ apply $A\_i^{-1}$ (forward/backward substitutions) + `prfe#update` --- ## [Step 5: Solve the linear system with preconditioned GMRES](../../documentation/ffddm/documentation.html#solving-the-linear-system) ```cpp func K[int] pr#fGMRES(K[int]& x0i, K[int]& bi, real eps, int itmax, string sp) ``` solves the linear system with flexible GMRES with DD preconditioner $M^{-1}$ - **x0i**: initial guess - **bi**: right-hand side - **eps**: relative tolerance - **itmax**: maximum number of iterations - **sp**: `"left"` or `"right"` preconditioning .pull-left[ .center[*left preconditioning*] .center[solve $M^{-1} A x = M^{-1} b$] ] .pull-right[ .center[*right preconditioning*] .center[solve $A M^{-1} y = b$] .center[$\Rightarrow x = M^{-1} y$] ] --- ```cpp macro dimension 2// EOM // 2D or 3D include "ffddm.idp" mesh ThGlobal = square(100,100); // global mesh // Step 1: Decompose the mesh ffddmbuildDmesh( M , ThGlobal , mpiCommWorld ) // Step 2: Define your finite element macro def(u) u // EOM macro init(u) u // EOM ffddmbuildDfespace( FE , M , real , def , init , P2 ) // Step 3: Define your problem macro grad(u) [dx(u), dy(u)] // EOM macro Varf(varfName, meshName, VhName) varf varfName(u,v) = int2d(meshName)(grad(u)'* grad(v)) + int2d(meshName)(1*v) + on(1, u = 0); // EOM ffddmsetupOperator( PB , FE , Varf ) FEVhi ui, bi; ffddmbuildrhs( PB , Varf , bi[] ) *// Step 4: Define the one level DD preconditioner *ffddmsetupPrecond( PB , Varf ) *// Step 5: Solve the linear system with GMRES *FEVhi x0i = 0; *ui[] = PBfGMRES(x0i[], bi[], 1.e-6, 200, "right"); ``` ```cpp ffddmplot(FE, ui, "u") PBwritesummary ``` --- ## [Define a two level DD preconditioner](../../documentation/ffddm/documentation.html#two-level-preconditioners) **Goal** improve scalability of the one level method $\Rightarrow$ enrich the one level preconditioner with a *coarse problem* coupling all subdomains Main ingredient is a rectangular matrix $\color{red}{Z}$ of size $n \times n\_c,\,$ where $n\_c \ll n$ $\color{red}{Z}$ is the *coarse space* matrix The *coarse space operator* $E = \color{red}{Z}^T A \color{red}{Z}$ is a square matrix of size $n\_c \times n\_c$ The simplest way to enrich the one level preconditioner is through the *additive coarse correction* formula: $$M^{-1}\_2 = M^{-1}\_1 + \color{red}{Z} E^{-1} \color{red}{Z}^T$$ .center[*How to choose $\color{red}{Z}$ ?*] --- ## [Build the GenEO coarse space](../../documentation/ffddm/documentation.html#building-the-geneo-coarse-space) ```cpp ffddmgeneosetup( pr , Varf ) ``` The *GenEO* method builds a robust coarse space for highly heterogeneous or anisotropic **SPD** problems $\Rightarrow$ solve a local generalized eigenvalue problem in each subdomain $$ D\_i A\_i D\_i\, V\_{i,k} = \lambda\_{i,k}\, A\_i^{\text{Neu}} \,V\_{i,k} $$ with $A\_i^{\text{Neu}}$ the local Neumann matrices built from **Varf** (same **Varf** as [Step 3](tutorial-slides.html#11)) The GenEO coarse space is $\color{red}{Z} = (R\_i^T D\_i V\_{i,k})^{i=1,...,N}\_{\lambda\_{i,k} \ge \color{blue}{\tau}}$ The eigenvectors $V\_{i,k}$ selected to enter the coarse space correspond to eigenvalues $\lambda_{i,k} \ge \color{blue}{\tau}$, where $\color{blue}{\tau}$ is a threshold parameter **Theorem** the spectrum of the preconditioned operator lies in the interval $[\displaystyle \frac{1}{1+k_1 \color{blue}{\tau}} , k_0 ]$ where $k_0 - 1$ is the \# of neighbors and $k_1$ is the multiplicity of intersections $\Rightarrow$ $k_0$ and $k_1$ do not depend on $N$ nor on the PDE --- ```cpp macro dimension 2// EOM // 2D or 3D include "ffddm.idp" mesh ThGlobal = square(100,100); // global mesh // Step 1: Decompose the mesh ffddmbuildDmesh( M , ThGlobal , mpiCommWorld ) // Step 2: Define your finite element macro def(u) u // EOM macro init(u) u // EOM ffddmbuildDfespace( FE , M , real , def , init , P2 ) // Step 3: Define your problem macro grad(u) [dx(u), dy(u)] // EOM macro Varf(varfName, meshName, VhName) varf varfName(u,v) = int2d(meshName)(grad(u)'* grad(v)) + int2d(meshName)(1*v) + on(1, u = 0); // EOM ffddmsetupOperator( PB , FE , Varf ) FEVhi ui, bi; ffddmbuildrhs( PB , Varf , bi[] ) // Step 4: Define the one level DD preconditioner ffddmsetupPrecond( PB , Varf ) *// Build the GenEO coarse space *ffddmgeneosetup( PB , Varf ) // Step 5: Solve the linear system with GMRES FEVhi x0i = 0; ui[] = PBfGMRES(x0i[], bi[], 1.e-6, 200, "right"); ``` --- ## [Build the coarse space from a coarse mesh](../../documentation/ffddm/documentation.html#building-the-coarse-space-from-a-coarse-mesh) ```cpp ffddmcoarsemeshsetup( pr , Thc , VarfEprec , VarfAprec ) ``` For **non SPD** problems, an alternative is to build the coarse space by discretizing the PDE on a coarser mesh **Thc** .red[$Z$] will be the *interpolation matrix* from the coarse FE space ${Vh}\_c$ to the original FE space $Vh$ $\Rightarrow E=\color{red}{Z}^{T} A \color{red}{Z}$ is the matrix of the problem discretized on the coarse mesh The variational problem to be discretized on **Thc** is given by macro **VarfEprec** **VarfEprec** can differ from the original **Varf** of the problem *Example*: added absorption for wave propagation problems Similarly, **VarfAprec** specifies the global operator involved in multiplicative coarse correction formulas. It defaults to $A$ if **VarfAprec** is not defined --- ```cpp macro dimension 2// EOM // 2D or 3D include "ffddm.idp" mesh ThGlobal = square(100,100); // global mesh // Step 1: Decompose the mesh ffddmbuildDmesh( M , ThGlobal , mpiCommWorld ) // Step 2: Define your finite element macro def(u) u // EOM macro init(u) u // EOM ffddmbuildDfespace( FE , M , real , def , init , P2 ) // Step 3: Define your problem macro grad(u) [dx(u), dy(u)] // EOM macro Varf(varfName, meshName, VhName) varf varfName(u,v) = int2d(meshName)(grad(u)'* grad(v)) + int2d(meshName)(1*v) + on(1, u = 0); // EOM ffddmsetupOperator( PB , FE , Varf ) FEVhi ui, bi; ffddmbuildrhs( PB , Varf , bi[] ) // Step 4: Define the one level DD preconditioner ffddmsetupPrecond( PB , Varf ) *// Build the coarse space from a coarse mesh *mesh Thc = square(10,10); *ffddmcoarsemeshsetup( PB , Thc , Varf , null ) // Step 5: Solve the linear system with GMRES FEVhi x0i = 0; ui[] = PBfGMRES(x0i[], bi[], 1.e-6, 200, "right"); ``` --- ## [Use **HPDDM** within **ffddm**](../../documentation/ffddm/documentation.html#using-hpddm-within-ffddm) **ffddm** allows you to use **HPDDM** to solve your problem, effectively replacing the **ffddm** implementation of all parallel linear algebra computations $\Rightarrow$ define your problem with **ffddm**, solve it with **HPDDM** $\Rightarrow$ **ffddm** acts as a finite element interface for **HPDDM** You can use **HPDDM** features unavailable in **ffddm** such as advanced Krylov subspace methods implementing block and recycling techniques To switch to **HPDDM**, simply define the macro `pr#withhpddm` before using `ffddmsetupOperator` ([Step 3](tutorial-slides.html#11)). You can then pass **HPDDM** options with command-line arguments or directly to the underlying **HPDDM** operator. Options need to be prefixed by the operator prefix: ```cpp macro PBwithhpddm()1 // EOM ffddmsetupOperator( PB , FE , Varf ) set(PBhpddmOP,sparams="-hpddm_PB_krylov_method gcrodr -hpddm_PB_recycle 10"); ``` Or, define `pr#withhpddmkrylov` to use **HPDDM** only for the Krylov method Example [here](https://github.com/FreeFem/FreeFem-sources/blob/develop/examples/ffddm/Helmholtz-2d-HPDDM-BGMRES.edp): Helmholtz problem with multiple rhs solved with Block GMRES --- ## Some results: Heterogeneous 3D elasticity with GenEO Heterogeneous 3D linear elasticity equation discretized with P2 FE solved on 4096 MPI processes $n\approx$ 262 million 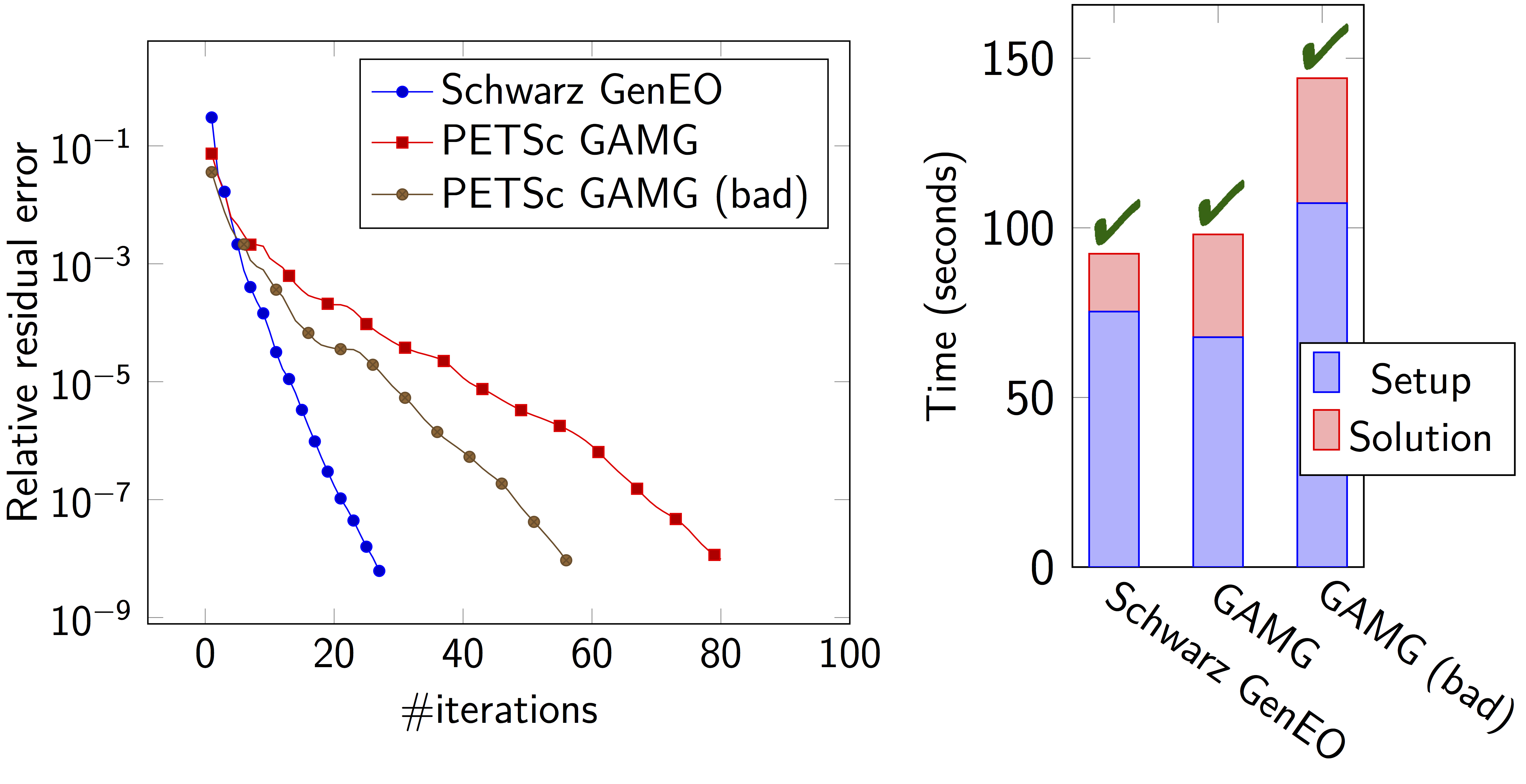 --- ## Some results: 2-level DD for Maxwell equations, scattering from the COBRA cavity <div style="overflow: hidden; display: inline-block;"> <div style="float:left; margin-left: auto; margin-right: auto;"> f = 10 GHz </div> <img src="../../_images/10GHz_borders_new_v2.png" width = 37% style="float; margin-left: 2%; margin-right: auto;" /> <img src="../../_images/10GHz_new.png" width = 45.7% style="float:right; margin-left: auto; margin-right: auto;"/> </div> <div style="overflow: hidden; display: inline-block;"> <div style="float:left; margin-left: auto; margin-right: auto;"> f = 16 GHz </div> <img src="../../_images/16GHz_new.png" width = 56% style="display:block; margin-left: auto; margin-right: auto;"/> </div> --- ## Some results: 2-level DD for Maxwell equations, scattering from the COBRA cavity - order 2 Nedelec edge FE - fine mesh: 10 points per wavelength - coarse mesh: 3.33 points per wavelength - two level ORAS preconditioner with added absorption - f = 10 GHz: $n\approx$ 107 million, .red[$n_c \approx$ 4 million] f = 16 GHz: $n\approx$ 198 million, .red[$n_c \approx$ 7.4 million] $\rightarrow$ coarse problem too large for a direct solver $\Rightarrow$ inexact coarse solve: GMRES + one level ORAS preconditioner 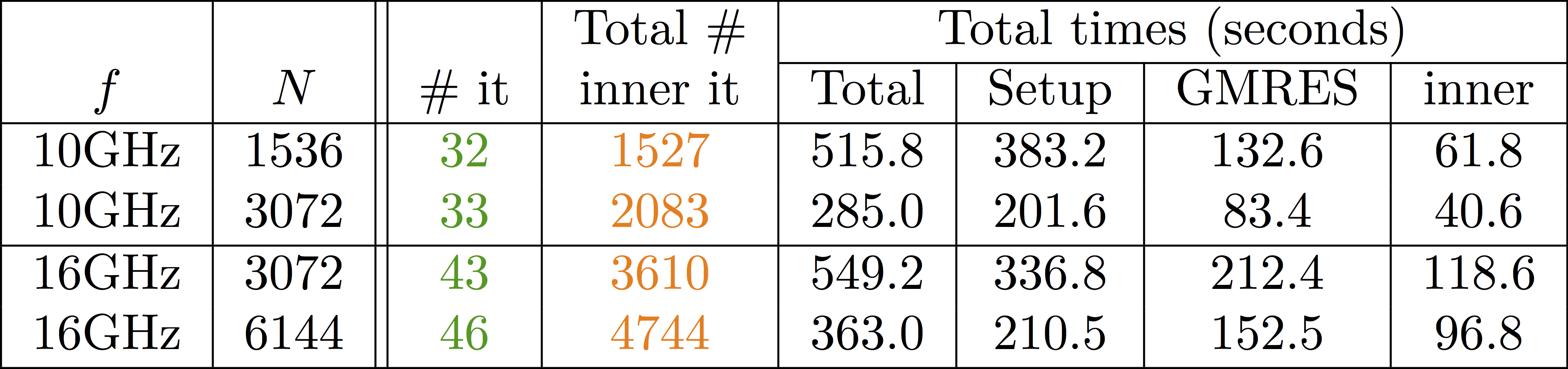 speedup of 1.81 from 1536 to 3072 cores at 10GHz 1.51 from 3072 to 6144 cores at 16GHz You can find the script [here](https://github.com/FreeFem/FreeFem-sources/blob/develop/examples/ffddm/Maxwell_Cobracavity.edp)